Towards Integrated Computational Biophysics and Engineering of Biomolecules
Probing Nucleic Acid Dynamics with Low-Resolution Data and Data-driven Simulations
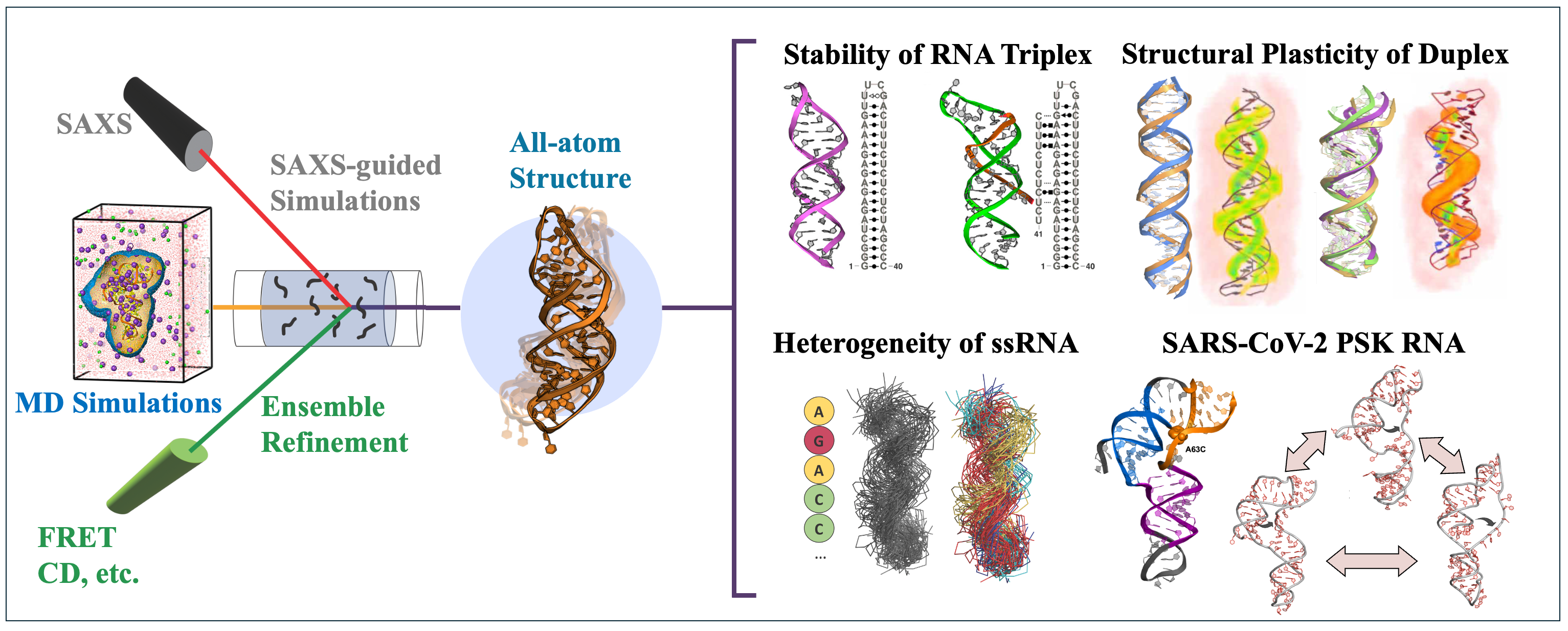
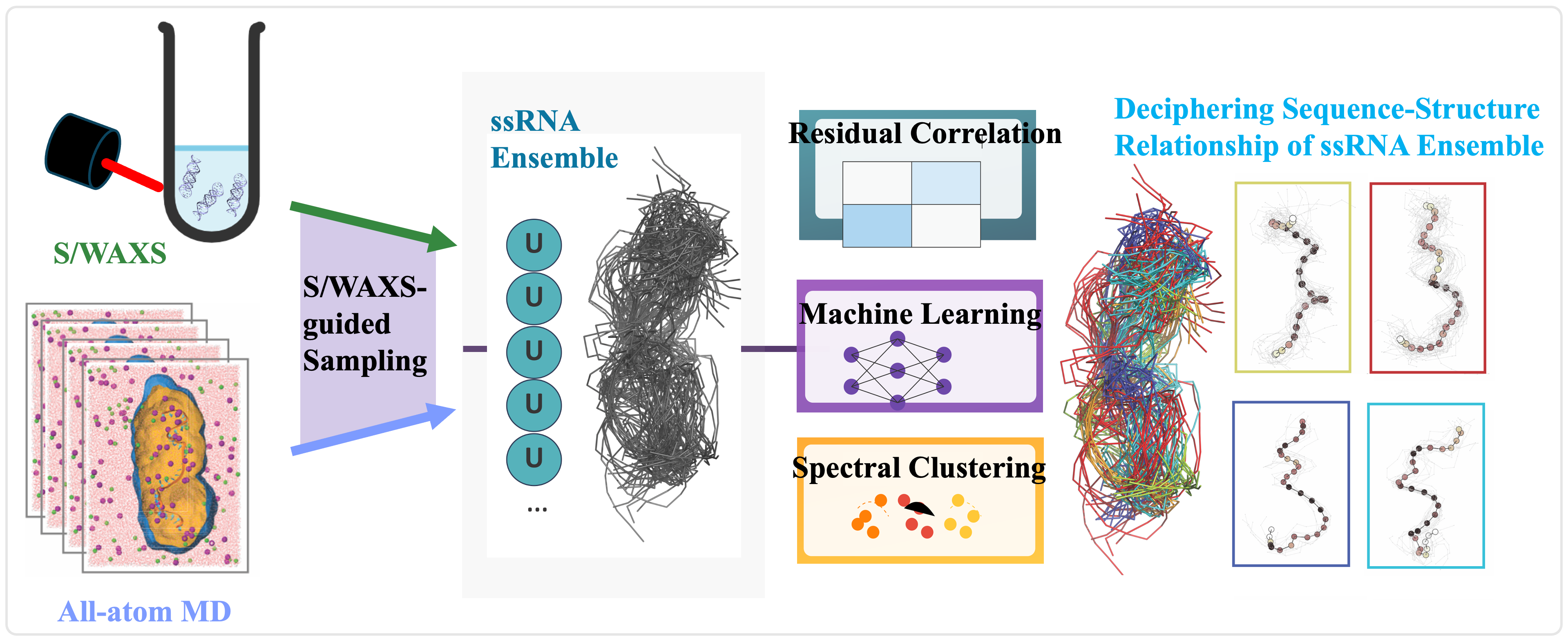
Probing the dynamic structure of nucleic acids remains a fundamental challenge due to their conformational flexibility and the limitations of experimental resolution. While solution SAXS provides valuable global shape information, its low resolution makes it difficult to resolve structural details essential for mechanistic insights. I mainly focused on bridging this gap through the integratiion of SAXS with molecular simulations to achieve atomistic interpretations of DNA and RNA dynamics. I focus on three key areas: enhanced sampling, experimental data-driven MD, and ensemble refinement. This approach has enabled the resolution of complex nucleic acid structures in solution and the discovery of new regulatory mechanisms in RNA biology.
Refining RNA Force Fields with Experimental Integration
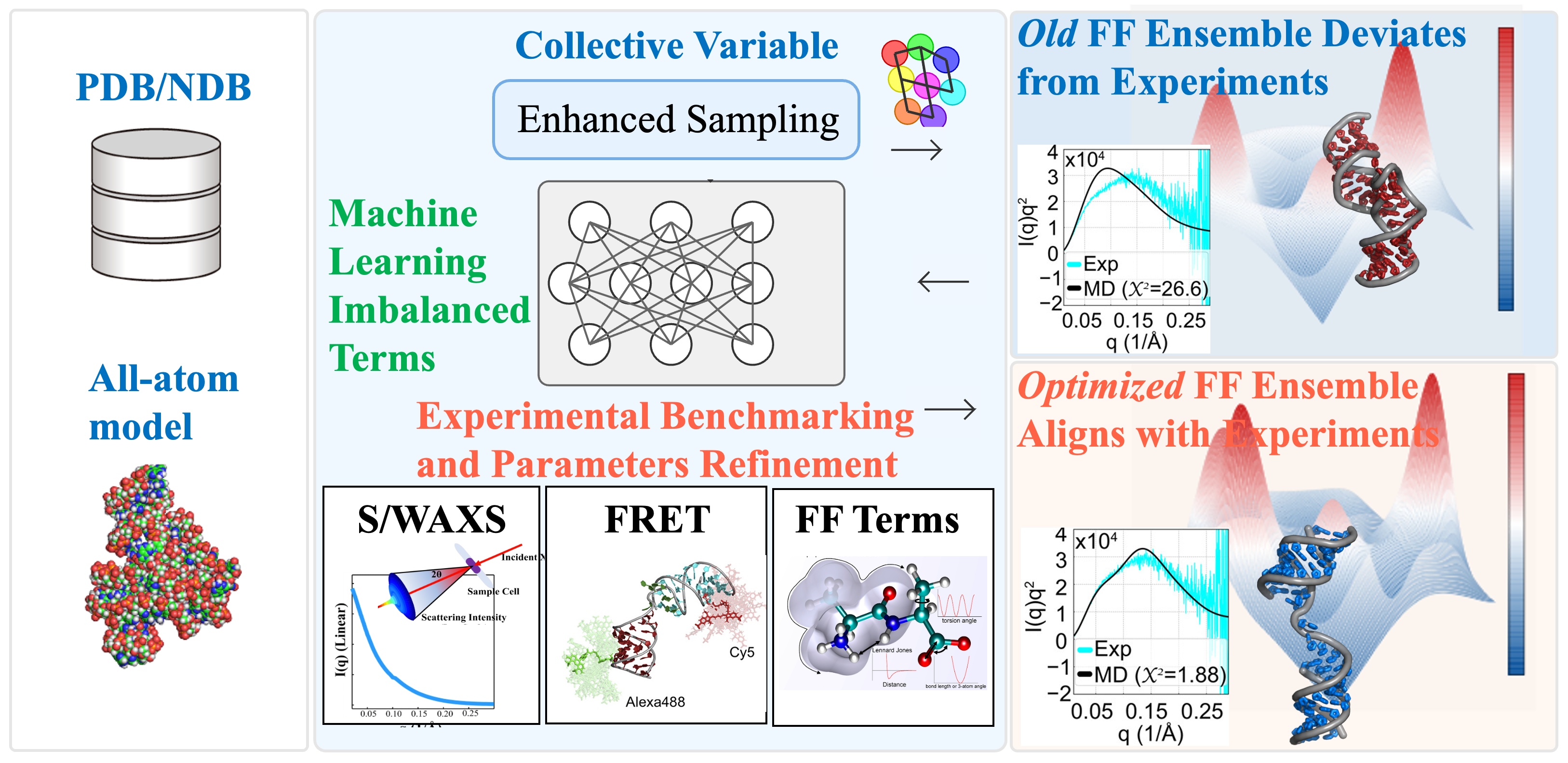
Accurately modeling RNA remains a major challenge due to its flexible backbone and complex energy landscape, which are often inadequately described by existing emprical force fields. To address this, my research focuses on refining RNA force field parameters by integrating MD simulations with experimental benchmarks from solution experiemnts, such as SAXS/WAXS and FRET. I applied machine learning methods to analyze simulation deviations from experimental data, to identify imbalances in energy terms—such as electrostatics and van der Waals interactions. This informed a targeted refinement strategy, resulting in a force field that significantly improves MD accuracy for flexible RNA motifs like single-stranded and Helix-Junction-Helix (HJH) structures. The refined model achieves enhanced agreement with experimental observables and provides a more faithful representation of RNA dynamics in solution.
Understanding Nucleic Acid Assembly in Non-Viral Gene Delivery Systems
Gene therapy relies on the efficient encapsulation and delivery of nucleic acid drugs, such as DNA and RNA, into target cells. The ability of these molecules to assemble and condense within delivery vectors is essential for therapeutic efficacy. My research focuses on the physical principles underlying nucleic acid–vector interactions, particularly in non-viral systems using cationic lipid nanoparticles (LNPs). I developed a theoretical and simulation-based framework to probe how DNA assembles and disassembles under varying conditions. This approach has been applied to study the self-organization of DNA–cationic lipid complexes, shedding light on their structural dynamics and informing the rational design of synthetic, low-toxicity delivery vectors for RNA therapeutics and mRNA vaccines.

